Ribosome stalling by SecM
The peptide SecM exists solely to stall the ribosome synthesizing it. SecM has an intrinsic stalling capability. Stalling of the ribosome synthesizing SecM provides time for a downstream RNA helix on the same mRNA strand to unwind. Unwinding of this helix then allows for a new ribosome to bind and synthesize a new protein, SecA, a bacterial ATP-driven translocase that aids the passage of nascent proteins across membranes in conjunction with SecY (see also our translocon research page). When sufficient levels of SecA have been reached, SecA interacts with the SecM-stalled ribosome to pull on SecM, freeing it and allowing translation to resume (illustrated schematically in the figure below). SecM, which serves no other purpose than to stall the ribosome, is released into the cell and degraded.
Much of what is known about SecM stalling comes from biochemical experiments, in which every amino acid in SecM’s sequence has been mutated and the resulting effects measured. These experiments revealed few residues as critical, although one stands out in every species as invariable: arginine 163 (R163, see the figure). However, until recently, little was known about the precise mechanisms at work. A cryo-EM map of a SecM-stalled ribosome revealed a shifted linkage in the PTC between the P-site tRNA and the SecM peptide. Although the shift was only 0.2 nm, it was hypothesized to be sufficient to inhibit peptide-bond formation, preventing synthesis of the remainder of SecM.
In order to elucidate precisely how SecM stalls the ribosome, we first applied MDFF to the cryo-EM map of the SecM-stalled ribosome to develop an atomic model of the complex. Next, we carried out extensive simulations of this wild-type model along with mutants of both the ribosome and SecM known to disrupt stalling. We discovered a putative communication pathway between the critical R163 and the PTC that causes the observed shifted linkage; the pathway is shown schematically in the figure. Finally, through steered MD simulations in which we pulled on the N-terminal end of SecM outside the ribosome, we determined that the pulling action of SecA can break the crucial interactions identified in our previous simulations and, thus, alleviate stalling.
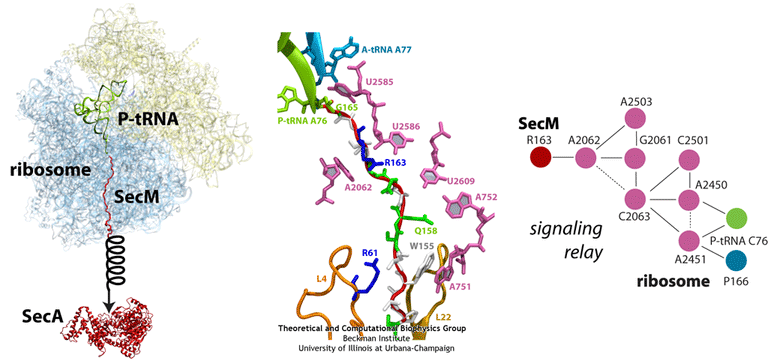
(left) Structure of the SecM-stalled ribosome determined with MDFF. SecA is shown for visualization purposes only. (middle) Atomic-scale view of SecM in the ribosome’s exit tunnel, near the PTC at the top. Residues involved in ribosome-SecM interactions are labeled. (right) Schematic of the communication pathway connecting SecM to the ribosome’s PTC.
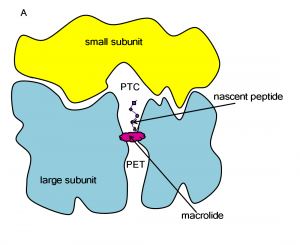
Macrolide-ribosome interactions
The ribosome is a large molecular machine, consisting of rRNA and proteins, that is responsible for protein synthesis in all cells. During protein synthesis, the genetic information in mRNA is decoded into a sequence of amino acids and these amino acids are linked together with peptide bonds, one at a time. Both the recognition and linking of amino acids occur in the peptidyl transferase center (PTC), while the nascent peptide exits the ribosome through the polypeptide exit tunnel (PET). Since there are significant structural differences between eukaryotic and prokaryotic ribosomes, the ribosome makes an excellent target for antibiotics, which can kill harmful bacteria without harming the host eukaryotic cells.
One class of antibiotics that targets the bacterial ribosome is macrolides and their derivatives. Their general structure is a 14-, 15- or 16-atom cyclic lactone ring with attached sugars, peptides or other functional groups.
Macrolides operate by binding in the ribosome’s PET, blocking the exit of nascent peptides. The blockage prevents continued synthesis once the nascent peptide reaches the macrolide, and the unfinished peptide dissociates from the ribosome. Unfortunately, due to the extensive use of macrolides, several strains of bacteria have become resistant to them. According to experimental studies, exchange or methylation of specific rRNA residues in the ribosome induces resistance to some or all macrolides.
We have used MD simulations to understand how specific rRNA mutations result in macrolide resistance and to facilitate the development of compounds that can overcome this resistance. We have simulated several macrolides in both wild-type and macrolide-resistant ribosomes. It was found that modifications in the resistant ribosomes led to less favorable interactions with the macrolides, displacing them from their crystal-structure position. We also studied several novel macrolides with additional indole-analog aromatic moieties. These moieties were added in order to overcome macrolide resistance by mimicking W155 and A751 interactions between SecM and the ribosome. Surprisingly, we found that the different indole-analog moieties could adopt varied geometries when interacting with A751, explaining the differences in the observed experimental activities.
Folding in the ribosome’s PET
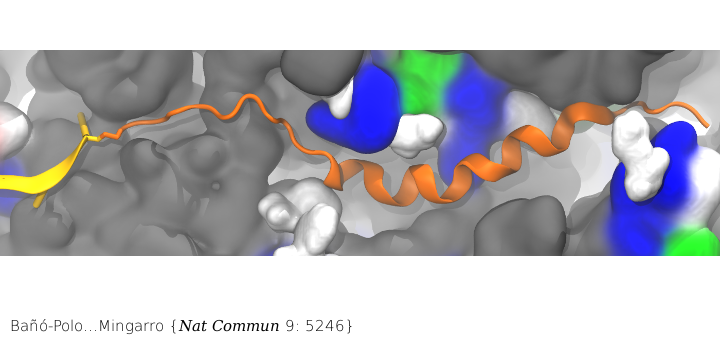
Nascent peptide within the PET. The nascent peptide is shown in cartoon representation (orange) bound to tRNA (yellow). Ribosomal proteins are shown in a surface representation and colored by residue type: (white) hydrophobic, (green) polar, and (blue) positively charged. Ribosomal RNA is also shown in a surface representation and colored gray.
Surprisingly, membrane proteins can begin folding even when they are still in the ribosome’s PET. While co-translational folding of nascent chains into an α-helical conformation in the ribosomal tunnel has been demonstrated previously, the features governing this folding are not well understood. In particular, little is known about what features influence the propensity to acquire α-helical structure in the ribosome. Combining our MD simulations with in vitro translation of truncated nascent chains trapped within the ribosome tunnel in the lab of Ismael Mingarro, we demonstrated that folding in the ribosome is attained for transmembrane helices but not for soluble helices, presumably facilitating SRP (signal recognition particle) recognition and/or a favorable conformation for membrane integration upon translocon entry.
Related publications
- Transmembrane but not soluble helices fold inside the ribosome tunnel.
M. Bañó-Polo, C. Baeza-Delgado, S. Tamborero, A. Hazel, B. Grau, I. Nilsson, P. Whitley, J. C. Gumbart, G. von Heijne, and I Mingarro. Nature Communications. 9:5246, 2018. - Toward the rational design of macrolide antibiotics to combat resistance.
A. Pavlova, J. M. Parks, A. K. Oyelere, and J. C. Gumbart. Chemical Biology & Drug Design. 90:641-652, 2017. - Parametrization of macrolide antibiotics using the force field toolkit.
A. Pavlova and J. C. Gumbart. J. Computational Chemistry. 36:2052-2063, 2015. - Mechanisms of SecM-mediated stalling in the ribosome.
J. Gumbart , E. Schreiner, D. N. Wilson, R. Beckmann, and K. Schulten. Biophysical Journal, 103:331-341, 2012.