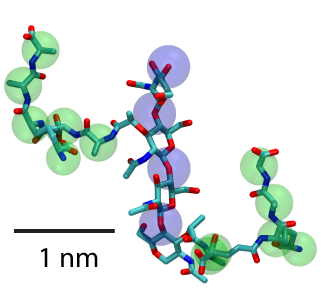
Two units of peptidoglycan with sugars highlighted by blue spheres and peptide cross-linkers by green.
Bacteria are under significant internal turgor pressure, due to high concentration of solutes inside the cell compared to outside. To protect themselves from rupturing, they possess a cell wall, a porous mesh-like structure composed of peptidoglycan. Peptidoglycan is a polymer made up of stiff glycan strands cross-linked by short, flexible peptides. While the chemical structure of the basic repeat unit is clear, how it is organized into a macroscopic wall is not. Two main proposals distinguished by the orientation of the glycan strands have been put forth. In one, the layered model, the glycan strands run circumferentially around the cell body; in the other scaffold model, they are perpendicular to the membrane. To test these models, we have constructed and simulated at an atomic scale patches of both Gram-positive and Gram-negative bacterial cell walls in different organizations up to 50 nanometers in size.
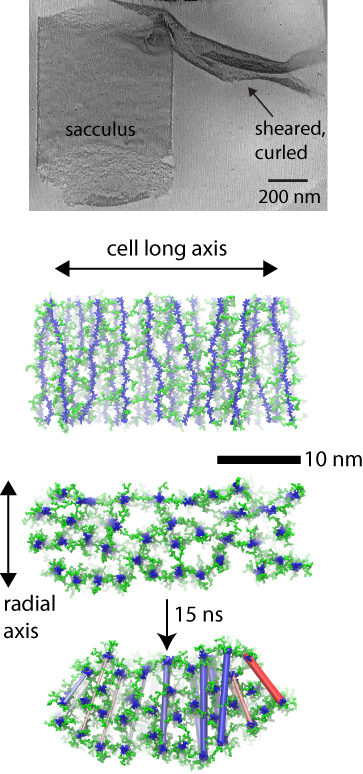
(top) cryo-tomography data. (middle) layered model simulated. (bottom) curling resulting in simulation.
Gram-positive cell wall simulations
In the case of the Gram-positive cell wall, the simulations elucidate the mechanisms behind a distinct curling effect observed in three-dimensional electron cryo-tomography images. Namely, when mechanically sheared, patches of the cell wall are observed to curl up on themselves in a direction orthogonal to their original curvature around the cell. Simulations of a layered model, but not of a scaffold model, recapitulate the curling behavior, which is found to arise due to a differential tension at the inside and outside of the patch.
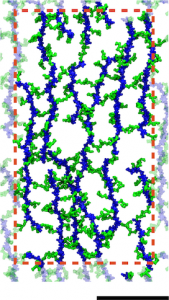
A model of the E. coli cell wall. The scale bar is 10 nm and the red dotted line indicates the unit cell in the simulations.
Gram-negative cell wall simulations
Gram-negative cell walls differ slightly in chemical composition from Gram-positive ones, but more importantly, they are only a single layer. Simulations were used to probe the mechanical properties of different arrangements of the cell wall, which were found to be in agreement with experiments. The simulations provided additional support for a disorganized, layered model of the cell wall. Furthermore, they demonstrate the connection between small-scale organization of a polymeric mesh of peptidoglycan and its cell-scale behavior.
Coarse-grained cell wall simulations
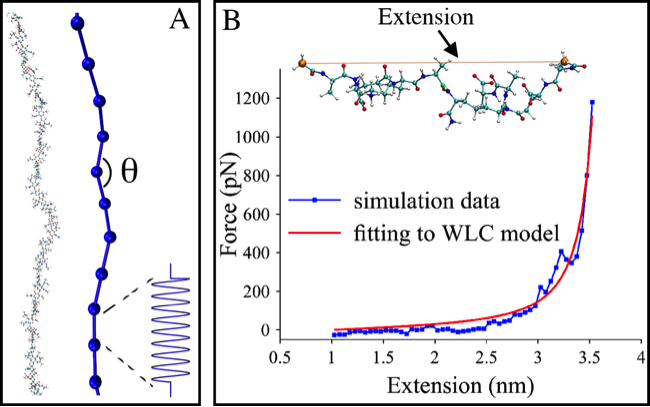
In collaboration with Lam Nguyen and Grant Jensen at Caltech, we have also investigated how peptidoglycan behaves on the scale of an entire cell using coarse-grained simulations developed by Lam. Based on the simulations, it was determined that sustainable growth of the cell wall can only occur when all enzymes involved in its remodeling are spatially and temporally coordinated. To parameterize the CG models, we used atomistic simulations of glycan chains and short peptides.
Coupling to the outer membrane
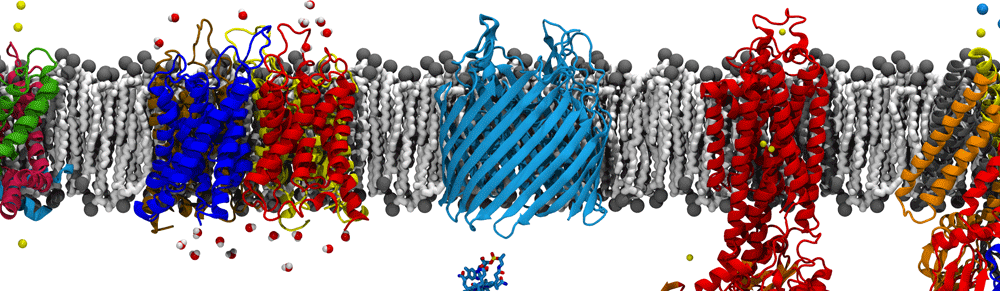
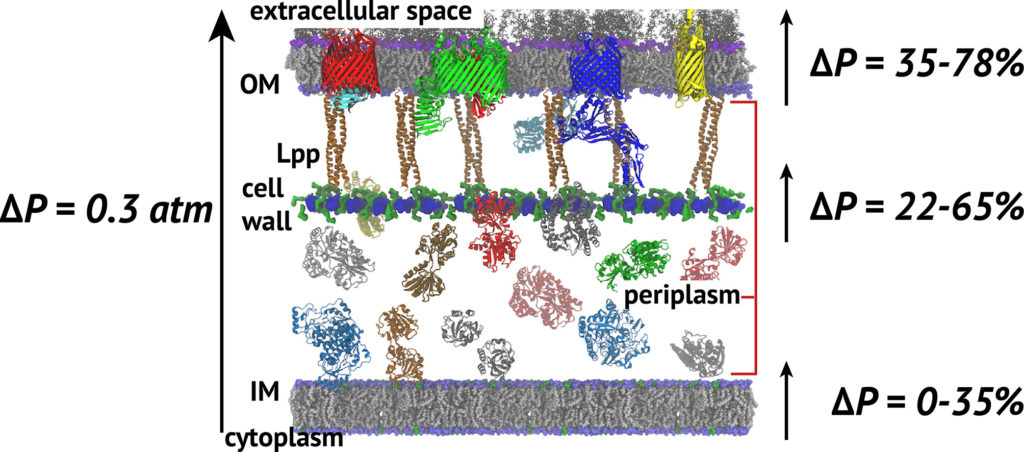
The cell wall sits between the inner and outer membranes (IM and OM) in Gram-negative bacteria but how it couples to them to share the stress generated through the turgor pressure is not well understood. In this study, we used MD simulations combined with experiments to resolve how and to what extent models of the IM, OM, and cell wall respond to changes in surface tension. We calculated the area compressibility modulus of all three components in simulations from tension-area isotherms. Experiments on monolayers mimicking individual leaflets of the IM and OM were also used to characterize their compressibility. While the membranes become softer as they expand, the cell wall exhibits significant strain stiffening at moderate to high tensions. We integrate these results into a model of the cell envelope in which the OM and cell wall share the tension at low turgor pressure (0.3 atm) but the tension in the cell wall dominates at high values (>1 atm).
Related publications
- Distribution of mechanical stress in the Escherichia coli cell envelope.
H. Hwang, N. Paracini, J. M. Parks, J. H. Lakey, and J. C. Gumbart. Biochimica et Biophysica Acta (BBA) – Biomembranes. 1860:2566-2575, 2018. - Coarse-grained simulations of bacterial cell wall growth reveal that local coordination alone can be sufficient to maintain rod shape. L. T. Nguyen, J. C. Gumbart, M. Beeby, and G. J. Jensen. Proceedings of the National Academy of Sciences, USA. 112:E3689–E3698, 2015
- Escherichia coli peptidoglycan structure and mechanics as predicted by atomic-scale simulations.
J. C. Gumbart, M. Beeby, G. Jensen, and B. Roux. PLOS Computational Biology, 10:e1003475, 2014. - Architecture and assembly of the Gram-positive cell wall.
M. Beeby, J. C. Gumbart, B. Roux, and G. J. Jensen. Mol. Microbiology, 88:664-672, 2013.