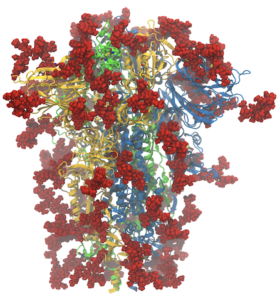
 Like many computational labs, at the start of the pandemic in the US in early 2020, we turned our attention to SARS-CoV-2, aided by a rapidly growing amount of structural data. Since then, as a community, we have learned a tremendous amount about the biophysics of the virus and the proteins it uses to infect and replicate. Our lab in particular has focused on protonation states of the main protease (Mpro), the closed-to-open transition of the spike protein, and its binding to the receptor ACE2 (see our coronavirus page for more). The last year and a half have also demonstrated a tremendous degree of communication and cooperation amongst the scientific community and especially the computational chemistry community, including in two ACS symposia at the spring meeting (“A Call to Action: The Many Roles of Computational Chemistry in Addressing COVID-19” organized by JC Gumbart and Rommie Amaro) and the fall meeting (“Computational Chemistry of COVID-19: Lessons Learned and Future Directions” organized by Katarzyna Świderek and Carlos Simmerling). What discoveries will the next year and a half bring?
Like many computational labs, at the start of the pandemic in the US in early 2020, we turned our attention to SARS-CoV-2, aided by a rapidly growing amount of structural data. Since then, as a community, we have learned a tremendous amount about the biophysics of the virus and the proteins it uses to infect and replicate. Our lab in particular has focused on protonation states of the main protease (Mpro), the closed-to-open transition of the spike protein, and its binding to the receptor ACE2 (see our coronavirus page for more). The last year and a half have also demonstrated a tremendous degree of communication and cooperation amongst the scientific community and especially the computational chemistry community, including in two ACS symposia at the spring meeting (“A Call to Action: The Many Roles of Computational Chemistry in Addressing COVID-19” organized by JC Gumbart and Rommie Amaro) and the fall meeting (“Computational Chemistry of COVID-19: Lessons Learned and Future Directions” organized by Katarzyna Świderek and Carlos Simmerling). What discoveries will the next year and a half bring?
SimBac – simulations of bacterial systems
…and other interesting biological phenomena