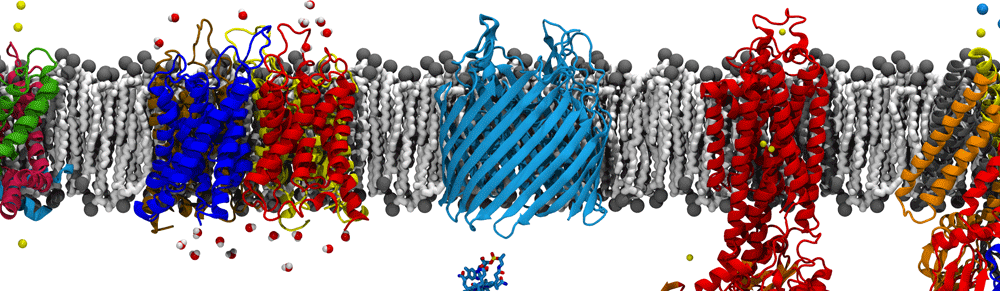
Visualizing Membrane Protein Insertion
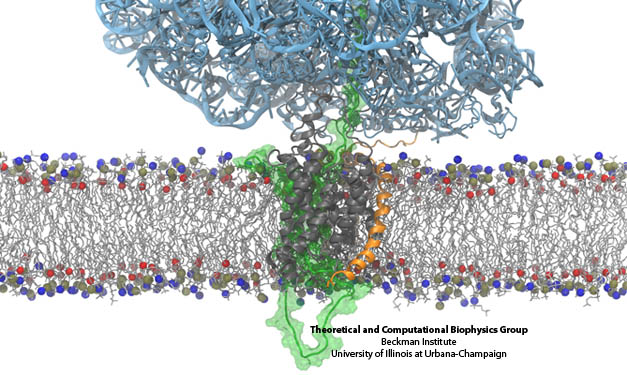
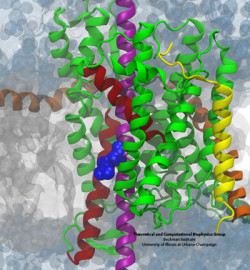
The derived ribosome-translocon-nascent-chain structure against an example lipid bilayer. The nascent protein (green) inserts into the channel where it loops back around to connect with the signal anchor at the translocon’s lateral gate.
Prior simulations investigating the membrane-insertion role of the translocon focused on the channel in the absence of an inserting protein. Evidence of the lateral gate’s role in membrane insertion of transmembrane segments has been largely indirect to date; for example, structures of a SecA-bound SecY and a Fab-bound SecY illustrated a slight opening of the gate, also in the absence of an inserting protein. A more recent structure of SecY with a symmetry-related copy inserting into the channel also exhibits modest gate opening. However, the insertion of a real nascent peptide remained unresolved. Just as happened previously (see the “translocon” research page), cryo-electron microscopy was used to capture a novel functional state of the translocon. In the experiments, carried out in the lab of Roland Beckmann at the University of Munich, a ribosome in the act of inserting a nascent protein into the channel was stalled and visualized by cryo EM. The channel was stabilized inside a self-contained lipid membrane, a so-called nanodisc, providing a native environment as opposed to the harsher detergents used to determine prior maps. Using the newly resolved map along with MDFF, we fitted atomic models of the ribosome, translocon, and nascent chain, revealing that the nascent protein’s N-terminal signal anchor was lodged in the translocon’s fully opened lateral gate, at the border between channel and membrane.
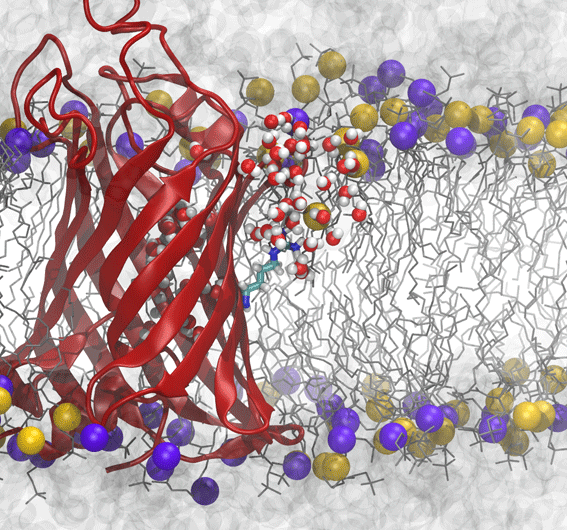
The MDFF-derived atomic structure was also used for free MD simulations containing the ribosome-translocon-nascent-chain complex as well as the complete nanodisc. In the simulations, an attraction between the lipids in the nanodisc and the ribosome’s helix 59 (H59) developed, with the lipids forming a small ridge. This ridge was found to be an excellent match with some otherwise inexplicable density in the original cryo-EM map, and led to the suggestion that H59 may play a role in aiding the inversion and subsequent insertion of the nascent chain’s signal anchor.
Quantifying Membrane Protein Insertion
Although we have visualized a trans-membrane segment, namely a signal anchor, in the process of inserting into the membrane, the structure does not address how the segment decides whether or not to insert. Systematic experiments testing the insertion propensity of various sequences of amino acids led to the suggestion that insertion is governed by purely by thermodynamics.
An unexpected result from those experiments was that charged residues such as arginine exhibit a relatively low penalty to be inserted into the membrane. Numerous computational and theoretical studies suggested a penalty of anywhere from 2 to 7 times what was observed experimentally, leading to confusion regarding what process was actually being measured.
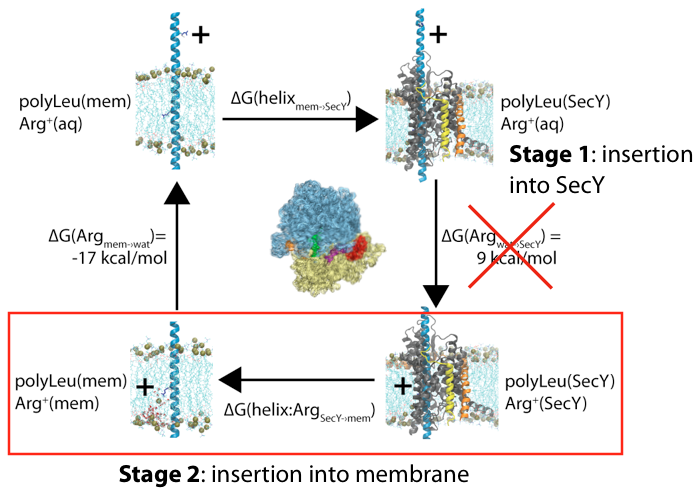
Membrane insertion via the ribosome-channel system can be decomposed into two stages, with only the energetics of the latter one being measured experimentally.
One potential source of the discrepancy between the experiments and simulations is in the assumption of the initial reference state. Although both presume that the end result is a transmembrane segment embedded in the bilayer, in the simulations the initial state was often chosen to be in a purely aqueous state, neglecting any potential role of the translocon. We hypothesized that the initial state for membrane insertion is, instead, the interior of the translocon. Using free-energy perturbation simulations, we tested this hypothesis by calculating the energetic cost to move an arginine and a leucine residue from the channel to the membrane. We found that the magnitude of both energies were less than in previous simulations, more closely matching the experimental results. Thus, protein insertion into the membrane likely follows a two-stage process, with the ribosome driving the protein into the channel in the first stage and then it exiting laterally through the gate in the second.
OmpLA – a challenge to the hypothesis
Although the two-stage model of insertion appeared to resolve a key discrepancy between simulations and experiments, new experimental results on the free-energy of insertion of the outer-membrane protein OmpLA once again complicated matters. OmpLA can insert spontaneously into lipid bilayers, without any translocon required and yet the insertion free energies were still quite low, similar to the biological scale. However, the bilayer used in experiments, DLPC, is much thinner than that used in previous simulations (DPPC). By computing the free-energy of arginine insertion on OmpLA in a DLPC bilayer, we found that the thin membrane explains the reduction in free energy cost for these new experiments.
Role of kinetics on insertion
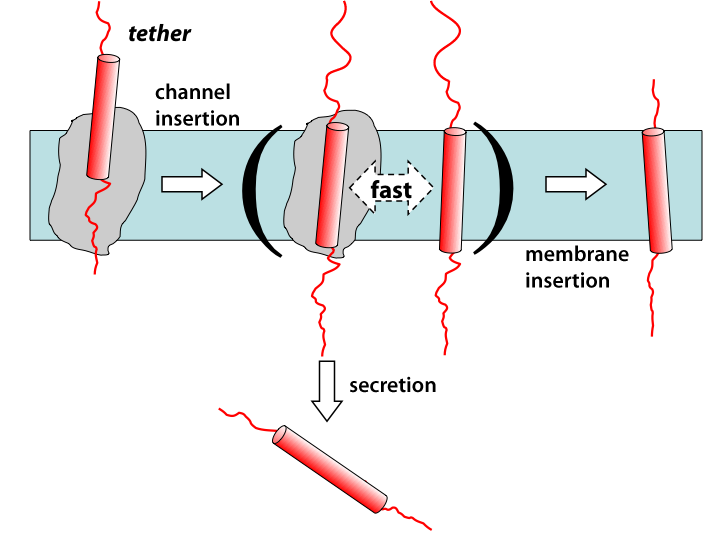
While it’s often been assumed that insertion from translocon to membrane occurs in equilibrium, including by the two-stage model described above, there are in fact many reasons to believe otherwise. A number of experiments over the years have shown that the insertion probability can be affected by apparent kinetic factors, e.g., the rate of translocation through the channel. To investigate such possibilities, we carried out simulations and free-energy calculations spanning time scales from microseconds (all-atom MD) up to seconds (finite-element diffusion). The simulations revealed that insertion could occur spontaneously, supporting the equilibrium picture, although the free energies were not in very good agreement with experimental numbers. However, the diffusion calculations revealed that when one takes into account that the inserting transmembrane segment is tethered to the ribosome by the rest of the slowly lengthening nascent chain, insertion probability becomes a function of both energetics and of time. This so-called “diffusion-elongation model” produced an apparent free-energy scale that looks like a two-state equilibrium one, but is compressed relative to the thermodynamic energies. Thus, it’s concluded that both kinetics and thermodynamics play roles in the insertion process.
Related publications
- Computed free energies of peptide insertion into bilayers are independent of computational method.
J. C. Gumbart, M. B. Ulmschneider, A. Hazel, S. H. White, and J. P. Ulmschneider. J. Membrane Biology. 251:345-356, 2018. - Decrypting protein insertion through the translocon with free-energy calculations.
J. C. Gumbart and C. Chipot. Biochimica et Biophysica Acta (BBA) – Biomembranes. 1858:1663–1671, 2016. - Reconciling the roles of kinetic and thermodynamic factors in membrane-protein insertion.
J. C. Gumbart, I. Teo, B. Roux, and K. Schulten. J. American Chemical Society, 135:2291-2297, 2013. - Determination of membrane-insertion free energies by molecular dynamics simulations.
J. Gumbart and B. Roux. Biophysical Journal, 102:795-801, 2012.
Featured in Biophysical Journal’s “Best of 2012”. - Free energy of nascent-chain folding in the translocon.
J. Gumbart, C. Chipot, and K. Schulten. J. American Chemical Society, 133:7602-7607, 2011. - Cryo-EM structure of the ribosome-SecYE complex in the membrane environment.
J. Frauenfeld, J. Gumbart, E. O. van der Sluis, S. Funes, M. Gartmann, B. Beatrix, T. Mielke, O. Berninghausen, T. Becker, K. Schulten, and R. Beckmann. Nature Structural & Molecular Biology, 18:614-621, 2011. - Free-energy cost for translocon-assisted insertion of membrane proteins.
J. Gumbart, C. Chipot, and K. Schulten. Proceedings of the National Academy of Sciences, USA, 108:3596-3601, 2011.