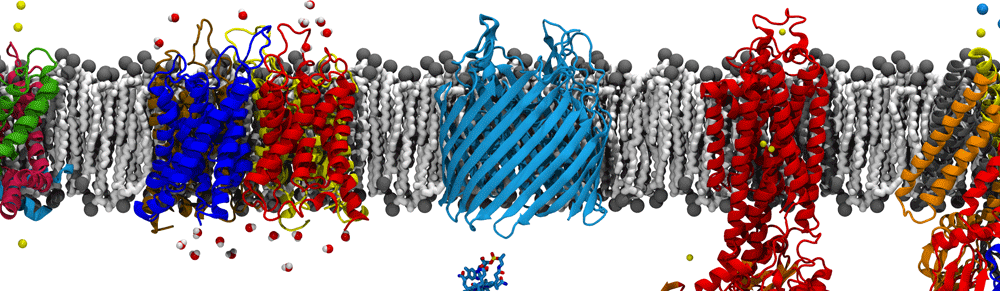
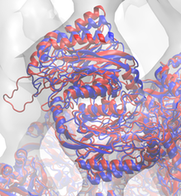
The molecular dynamics flexible fitting (MDFF) method can be used to flexibly fit atomic structures into density maps. The method consists of adding external forces proportional to the gradient of the density map into a molecular dynamics (MD) simulation of the atomic structure. Thus, high-resolution, but conformationally limited structures of individual components from, e.g., X-ray crystallography, can be fitted into cryo-electron microscopy maps of large, physiological complexes. I have utilized MDFF to determine the structures of a number of biologically and industrially relevant complexes, including ribosomes bound to the translocon (see above) and spiral-forming nitrilases. In the process, I have developed additional refinements to the MDFF method, such as incorporating symmetry-based restraints during fitting. I am also pushing the boundaries of MDFF by applying it to cryo-tomography data, which is typically at a significantly lower resolution than cryo-EM data.
MDFF is currently implemented in the MD package NAMD. More details can be found on the official MDFF webpage.
Publications
- Mechanisms of SecM-mediated stalling in the ribosome.
James Gumbart , Eduard Schreiner, Daniel N. Wilson, Roland Beckmann, and Klaus Schulten. Biophysical Journal, 103:331-341, 2012. - Symmetry-restrained flexible fitting for symmetric EM maps.
Kwok-Yan Chan, James Gumbart, Ryan McGreevy, Jean M. Watermeyer, B. Trevor Sewell, and Klaus Schulten. Structure, 19:1211-1218, 2011. - Cryo-EM structure of the ribosome-SecYE complex in the membrane environment.
Jens Frauenfeld, James Gumbart, Eli O. van der Sluis, Soledad Funes, Marco Gartmann, Birgitta Beatrix, Thorsten Mielke, Otto Berninghausen, Thomas Becker, Klaus Schulten, and Roland Beckmann. Nature Structural & Molecular Biology, 18:614-621, 2011. - Applications of the molecular dynamics flexible fitting method.
Leonardo G. Trabuco, Eduard Schreiner, James Gumbart, Jen Hsin, Elizabeth Villa, and Klaus Schulten. Journal of Structural Biology, 173:420-427, 2011. - Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome.
Thomas Becker, Shashi Bhushan, Alexander Jarasch, Jean-Paul Armache, Soledad Funes, Fabrice Jossinet, James Gumbart, Thorsten Mielke, Otto Berninghausen, Klaus Schulten, Eric Westhof, Reid Gilmore, Elisabet C. Mandon, and Roland Beckmann. Science, 326:1369-1373, 2009. - Regulation of the protein-conducting channel by a bound ribosome.
James Gumbart, Leonardo G. Trabuco, Eduard Schreiner, Elizabeth Villa, and Klaus Schulten. Structure, 17:1453-1464, 2009. - Protein-induced membrane curvature investigated through molecular dynamics flexible fitting.
Jen Hsin, James Gumbart, Leonardo G. Trabuco, Elizabeth Villa, Pu Qian, C. Neil Hunter, and Klaus Schulten. Biophysical Journal, 97:321-329, 2009.