The cell envelope in Gram-negative bacteria is made of two distinct membranes and a cell wall between them. There has been a growing interest in the mechanical adaptation of this cell envelope to the osmotic pressure (or turgor pressure), which is generated by the difference in the concentration of solute between the cytoplasm and the external environment. Via various measurement techniques, the turgor pressure values under physiological conditions have been estimated to be between 0.3 atm and 5 atm. It is, however, not clearly answered how the cell wall, the inner membrane (IM), and the outer membrane (OM) effectively protect the cell from the turgor pressure by sustaining the surface tension, which prevents the formation of inner membrane bulges, abnormal cell morphology, spheroplasts and cell lysis.
From a mechanics point of view, the IM maintains a higher concentration of solute in the cytoplasm than the external environment and the difference in osmotic pressure, namely the turgor pressure, pushes the membrane against the cell wall. The magnitude of the turgor pressure under physiological conditions has been estimated using several techniques including chemical and mechanical measurements, and the estimated pressure values vary by more than an order of magnitude, from 0.3 atm to 5 atm. It is clear that maintaining a proper turgor pressure is vital to cell viability, and the disruption of the cell wall will cause inner membrane bulges, abnormal cell morphology, spheroplasts or cell lysis. Unfortunately it is not yet answered how the surface tension generated by the turgor pressure is distributed between the cell wall and both membranes to give rise to overall protection against the turgor pressure. To resolve which membrane transmits stress from the turgor pressure to the cell wall, simulations of individual membranes as well as membranes coupled to the cell wall should be carried out, focusing on the effect of mechanical stress on membranes. Our simulations will utilize an applied surface tension to mimic the osmotic pressure effect as simulated in other MD simulation studies.

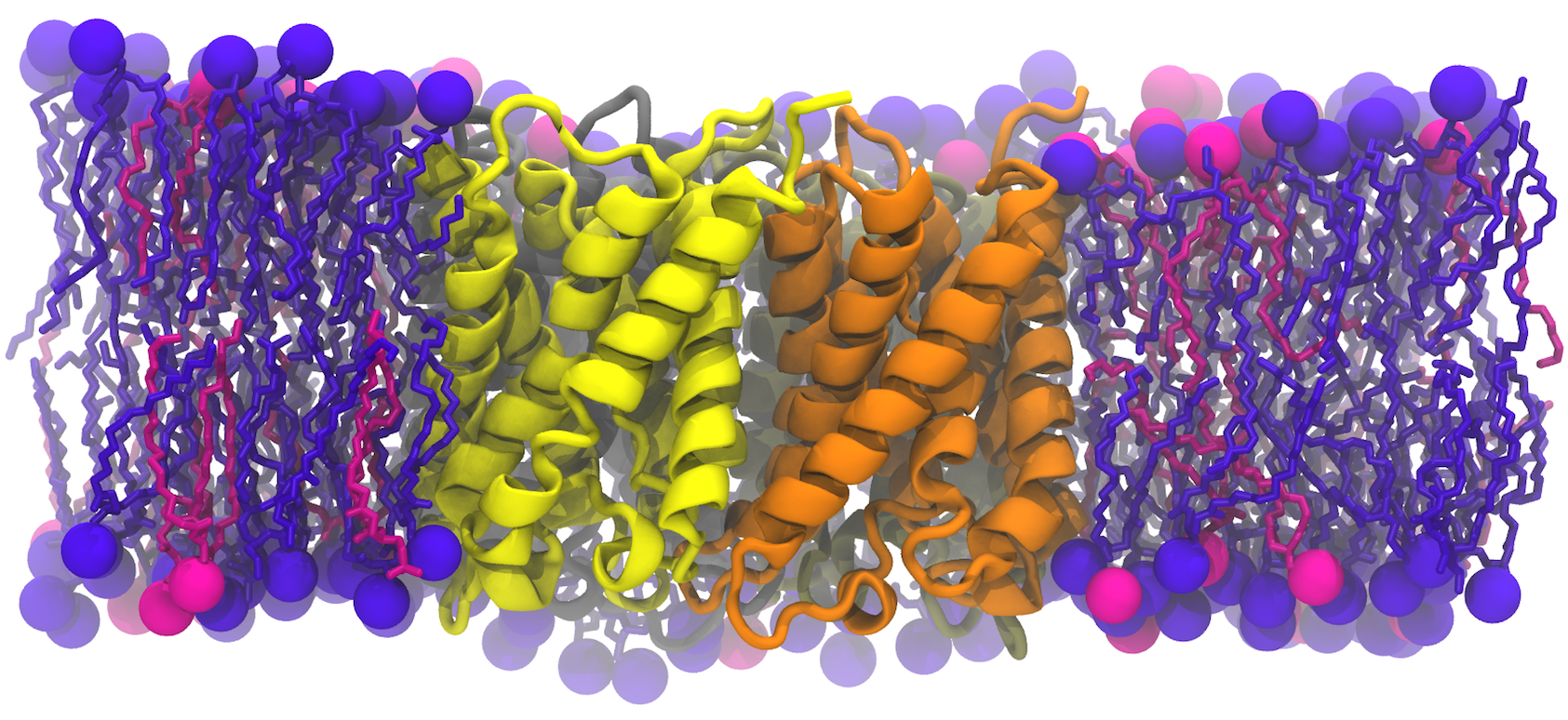

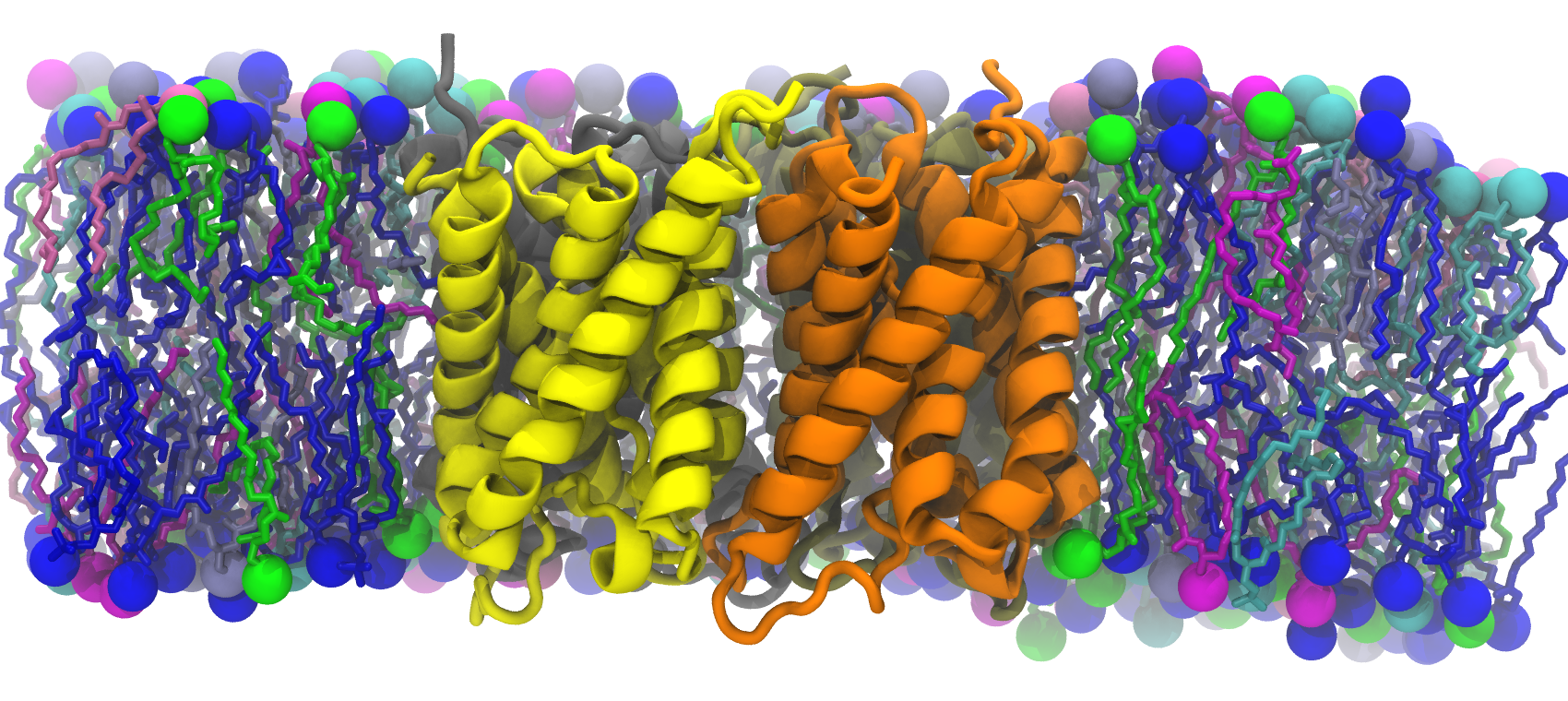
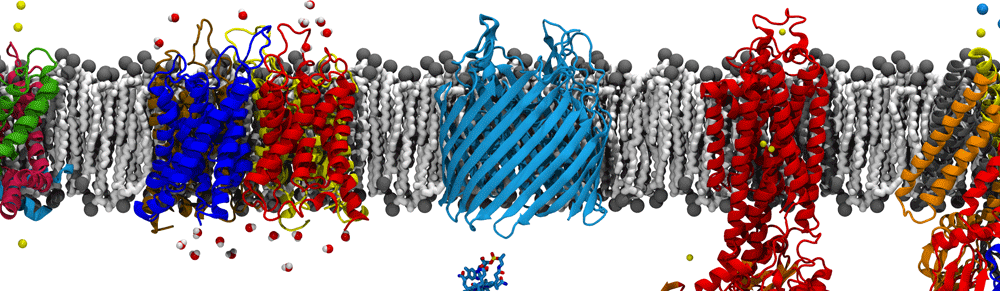
We used two models of the IM; the first membrane is modeled as a mixed 75%POPE/25% POPG bilayer while the second membrane model consists of saturated, unsaturated, and cycle-containing lipids that more accurately reflect the diverse population of lipids within the E. coli cytoplasmic membrane. Additionally, we built the bacterial OM, which has an outer leaflet of lipopolysaccharides that stiffen it. To rigorously mimic the aforementioned membrane systems, we have taken the presence of the membrane proteins into account as the models were prepared, based on previous studies that approximated 25% of protein contents in both membranes.
The outcomes of the simulation will be used to correlate the structural characteristics of the membranes and the mechanical resistance against the turgor pressure that is mimicked by applying surface tension as simulated in other MD simulation studies. In order to describe the elasticity with general mechanical properties, we focused on the low tension regime (elastic regime) which can be exploited to measure the elastic area compressibility modulus, the energy change, Young’s modulus, while non linear regime was also studied for cell wall to characterize its stress stiffening. Simulations of individual membranes as well as membranes coupled to the cell wall were also carried out.